Esmfold Folding Or Protein Structure Prediction

310.ai On LinkedIn: ESMFold: Folding Or Protein Structure Prediction
310.ai On LinkedIn: ESMFold: Folding Or Protein Structure Prediction Esm 2 outperforms all tested single sequence protein language models across a range of structure prediction tasks. esmfold harnesses the esm 2 language model to generate accurate structure predictions end to end directly from the sequence of a protein. We developed a new protein structure prediction approach named esmfold. esmfold uses the representations from a large language model (esm2) to generate an accurate structure prediction from the sequence of a protein.
GitHub - Jolenefu/ESMFold-Protein-Folding-demo
GitHub - Jolenefu/ESMFold-Protein-Folding-demo They created esmfold, a sequence to structure predictor that is nearly as accurate as alignment based methods and considerably faster. the increased speed permitted the generation of a database, the esm metagenomic atlas, containing more than 600 million metagenomic proteins. They created esmfold, a sequence to structure predictor that is nearly as accurate as alignment based methods and considerably faster. the increased speed permitted the generation of a database, the esm metagenomic atlas, containing more than 600 million metagenomic proteins. —maf. Esmfold (evolutionary scale modeling) is an artificial intelligence method for predicting protein structures. the method is described in: evolutionary scale prediction of atomic level protein structure with a language model. For short monomeric proteins under the length 400, consider using esmfold api (no need for gpu, super fast!).
GitHub - ArthrowAbstract/esmfold-protein-prediction: Building A Web App For Predicting The ...
GitHub - ArthrowAbstract/esmfold-protein-prediction: Building A Web App For Predicting The ... Esmfold (evolutionary scale modeling) is an artificial intelligence method for predicting protein structures. the method is described in: evolutionary scale prediction of atomic level protein structure with a language model. For short monomeric proteins under the length 400, consider using esmfold api (no need for gpu, super fast!). Esmfold is a state of the art end to end protein folding model based on an esm 2 backbone. it does not require any lookup or msa step, and therefore does not require any external databases to be present in order to make predictions. Esmfold – evolutionary scale modeling esmfold is a large language model approach to protein structure prediction by meta ai. unlike alphafold2, esmfold does not use sequence alignments during prediction. instead, esmfold learned to predict protein structures directly from sequence. Esmfold combines a frozen pretrained esm 2 language model with a specialized structure module to predict protein structures with high accuracy. Esmfold is a gpu accelerated protein structure prediction algorithm utilizing evolutionary scale protein language models to infer atomic level 3d coordinates directly from amino acid sequences, without requiring msas or templates.

Bioinformatics, Protein Structure Prediction, ESMFold, AlphaFold, Protein Function, Artificial ...
Bioinformatics, Protein Structure Prediction, ESMFold, AlphaFold, Protein Function, Artificial ... Esmfold is a state of the art end to end protein folding model based on an esm 2 backbone. it does not require any lookup or msa step, and therefore does not require any external databases to be present in order to make predictions. Esmfold – evolutionary scale modeling esmfold is a large language model approach to protein structure prediction by meta ai. unlike alphafold2, esmfold does not use sequence alignments during prediction. instead, esmfold learned to predict protein structures directly from sequence. Esmfold combines a frozen pretrained esm 2 language model with a specialized structure module to predict protein structures with high accuracy. Esmfold is a gpu accelerated protein structure prediction algorithm utilizing evolutionary scale protein language models to infer atomic level 3d coordinates directly from amino acid sequences, without requiring msas or templates. Esmfold is an ai tool that uses language models to predict protein structures quickly and accurately, rivaling other top ai tools and aiding scientific research. The esmfold online webserver allows anybody with a neurosnap account to run and access esmfold, no downloads required. information submitted through this webserver is kept confidential and never sold to third parties as detailed by our strong terms of service and privacy policy. This repository contains code and pre trained weights for transformer protein language models from facebook ai research, including our state of the art esm 2 and esmfold, as well as msa transformer, esm 1v for predicting variant effects and esm if1 for inverse folding. Esmfold: made by meta ai, esmfold is based on esm 2 (evolutionary scale modeling 2), a big protein language model using transformer. it predicts protein structure only from a single amino acid sequence, no need for evolutionary information (msa).

Decoding Protein Structures: From AlphaFold To Beyond
Decoding Protein Structures: From AlphaFold To Beyond Esmfold combines a frozen pretrained esm 2 language model with a specialized structure module to predict protein structures with high accuracy. Esmfold is a gpu accelerated protein structure prediction algorithm utilizing evolutionary scale protein language models to infer atomic level 3d coordinates directly from amino acid sequences, without requiring msas or templates. Esmfold is an ai tool that uses language models to predict protein structures quickly and accurately, rivaling other top ai tools and aiding scientific research. The esmfold online webserver allows anybody with a neurosnap account to run and access esmfold, no downloads required. information submitted through this webserver is kept confidential and never sold to third parties as detailed by our strong terms of service and privacy policy. This repository contains code and pre trained weights for transformer protein language models from facebook ai research, including our state of the art esm 2 and esmfold, as well as msa transformer, esm 1v for predicting variant effects and esm if1 for inverse folding. Esmfold: made by meta ai, esmfold is based on esm 2 (evolutionary scale modeling 2), a big protein language model using transformer. it predicts protein structure only from a single amino acid sequence, no need for evolutionary information (msa). We present esmfold for high accuracy end to end atomic level structure prediction directly from the individual sequence of a protein. esmfold has similar accuracy to alphafold2 and rosettafold for sequences with low perplexity that are well understood by the language model. Esmfold (evolutionary scale modeling) is an artificial intelligence method for predicting protein structures. the method is described in: evolutionary scale prediction of atomic level protein structure with a language model. Esmfold is a machine learning model that predicts protein 3d structures from amino acid sequences using transformer based neural networks. developed by meta (formerly facebook research), it leverages evolutionary data for fast and accurate predictions. Predicts the 3d structure of a protein from its amino acid sequence. esmfold is a protein structure prediction deep learning model developed by facebook ai research (fair) lin2023esmfold.

Using The Structure Prediction Tool — OpenProtein-Docs Documentation
Using The Structure Prediction Tool — OpenProtein-Docs Documentation Esmfold is an ai tool that uses language models to predict protein structures quickly and accurately, rivaling other top ai tools and aiding scientific research. The esmfold online webserver allows anybody with a neurosnap account to run and access esmfold, no downloads required. information submitted through this webserver is kept confidential and never sold to third parties as detailed by our strong terms of service and privacy policy. This repository contains code and pre trained weights for transformer protein language models from facebook ai research, including our state of the art esm 2 and esmfold, as well as msa transformer, esm 1v for predicting variant effects and esm if1 for inverse folding. Esmfold: made by meta ai, esmfold is based on esm 2 (evolutionary scale modeling 2), a big protein language model using transformer. it predicts protein structure only from a single amino acid sequence, no need for evolutionary information (msa). We present esmfold for high accuracy end to end atomic level structure prediction directly from the individual sequence of a protein. esmfold has similar accuracy to alphafold2 and rosettafold for sequences with low perplexity that are well understood by the language model. Esmfold (evolutionary scale modeling) is an artificial intelligence method for predicting protein structures. the method is described in: evolutionary scale prediction of atomic level protein structure with a language model. Esmfold is a machine learning model that predicts protein 3d structures from amino acid sequences using transformer based neural networks. developed by meta (formerly facebook research), it leverages evolutionary data for fast and accurate predictions. Predicts the 3d structure of a protein from its amino acid sequence. esmfold is a protein structure prediction deep learning model developed by facebook ai research (fair) lin2023esmfold. In this video we demonstrate how to perform protein folding / protein structure prediction using esmfold on neurosnap. You can predict a protein structure from a sequence using esmfold (evolutionary scale modeling) from chimerax (daily build from november 9, 2022 or newer, not in 1.5). chimerax uses the prediction server provided by the esm metagenomic atlas described in this article. Using data from known protein structures and sequences, scientists developed an artificial intelligence (ai) workflow to predict the structures and functions of unknown proteins, including how these proteins would interact with metals such as zinc. in this example, predicted to be a zinc binding protein, the model of the protein shows that four cysteine residues are directly involved in the.
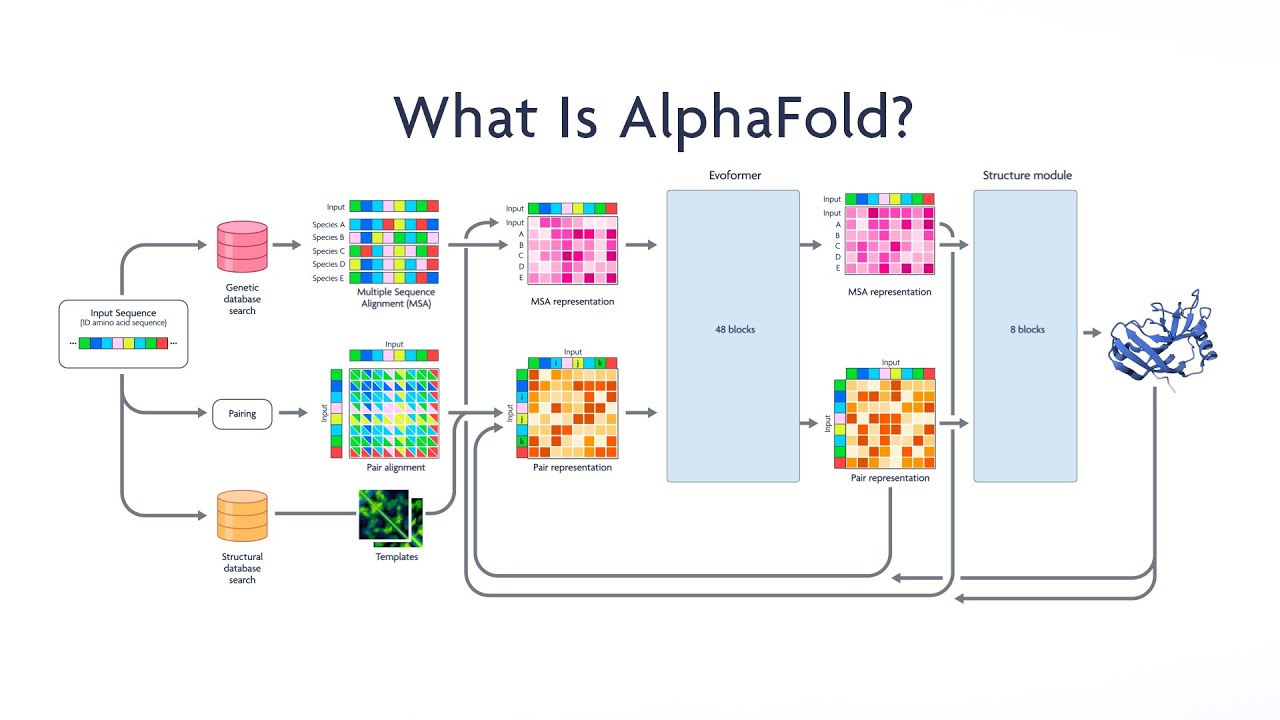
What Is AlphaFold? | NEJM
What Is AlphaFold? | NEJM
Related image with esmfold folding or protein structure prediction
Related image with esmfold folding or protein structure prediction
About "Esmfold Folding Or Protein Structure Prediction"














Comments are closed.